Identifying genetic mutation in glioma using multi-MRI radiomics and topological signatures
Identifying Genetic Mutation in Glioma Using Multi-mri Radiomics and Topological Signatures
Friday, April 21, 2023

Sufyan Ibrahim, MBBS (he/him/his)
Research Fellow
Mayo Clinic Rochester
Rochester, Minnesota, United States
ePoster Presenter(s)
Introduction: Eighty percent of malignant brain tumors are gliomas, which form in the brain and stem from glial cells. Noninvasive discovery of gene mutations from glioma is important for prognosis and treatment planning.
Methods: Our goal is to investigate the predictiveness of radiomics and topological features from multi-MRI for mutation status prediction and further compare different predictive models. MRI data (T1, T2, FLAIR) and clinical information for two hundred thirty-eight glioma (high grade glioma and low grade glioma) patients (165 patients: train; 76 patients: test) were obtained from The Cancer Imaging Achieve (TCIA), and gene expression information was accessed from cbioPortal. Radiomics and topological features were extracted from the whole tumor region. Tmap, minimum redundancy maximum relevance, and correlation-based feature elimination were assessed for feature selection before utilizing SVM and random forests for prediction.
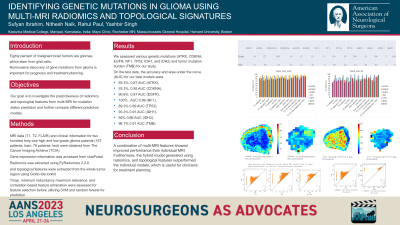
Results: We assessed various genetic mutations (ATRX, CDKNA, EGFR, NF1, TP53, IDH1, and IDH2) and tumor mutation burden (TMB) for our study. On the test data, the accuracy and area under the curve (AUC) for our best models were 93.3%, 0.87 AUC (ATRX), 93.3%, 0.95 AUC (CDKNA), 90.6% , 0.87 AUC (EGFR), 100% , AUC 0.99 (NF1), 89.3% 0.89 AUC (TP53), 93.3% 0.91 AUC (IDH1), 92% 0.96 AUC (IDH2), and 96.7% 0.97 AUC (TMB).
Conclusion : A combination of multi-MRI features showed improved performance than individual MRI. Furthermore, the hybrid model generated using radiomics, and topological features outperformed the individual models, which is useful for clinicians for treatment planning.
Methods: Our goal is to investigate the predictiveness of radiomics and topological features from multi-MRI for mutation status prediction and further compare different predictive models. MRI data (T1, T2, FLAIR) and clinical information for two hundred thirty-eight glioma (high grade glioma and low grade glioma) patients (165 patients: train; 76 patients: test) were obtained from The Cancer Imaging Achieve (TCIA), and gene expression information was accessed from cbioPortal. Radiomics and topological features were extracted from the whole tumor region. Tmap, minimum redundancy maximum relevance, and correlation-based feature elimination were assessed for feature selection before utilizing SVM and random forests for prediction.
Results: We assessed various genetic mutations (ATRX, CDKNA, EGFR, NF1, TP53, IDH1, and IDH2) and tumor mutation burden (TMB) for our study. On the test data, the accuracy and area under the curve (AUC) for our best models were 93.3%, 0.87 AUC (ATRX), 93.3%, 0.95 AUC (CDKNA), 90.6% , 0.87 AUC (EGFR), 100% , AUC 0.99 (NF1), 89.3% 0.89 AUC (TP53), 93.3% 0.91 AUC (IDH1), 92% 0.96 AUC (IDH2), and 96.7% 0.97 AUC (TMB).
Conclusion : A combination of multi-MRI features showed improved performance than individual MRI. Furthermore, the hybrid model generated using radiomics, and topological features outperformed the individual models, which is useful for clinicians for treatment planning.
